 CFR 610
CFR 610
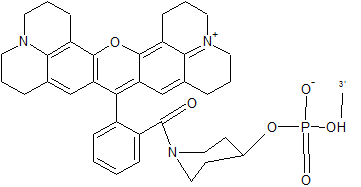
CFR 610 is an orange-red fluorophore. It is also used as a good alternative to fluorophore Texas Red. CFR 610 may be attached only to 5´end of an oligonucleotide.
The most common modifications are at the end of the string – at the 3’ or 5’ end, or combined. The modifying molecule may be in the middle of the chain, the bases in the chain may be replaced by their analogs; spacers of different lengths and nature, functional groups or molecules may be inserted into the chain. Modified phosphate linkage may be in the chain in all or only in selected positions. Less frequent modifications or their combinations are recommended to be consulted first because there are many limitations for synthesis and for purification of the resulting product.

CFR 610 is an orange-red fluorophore. It is also used as a good alternative to fluorophore Texas Red. CFR 610 may be attached only to 5´end of an oligonucleotide.
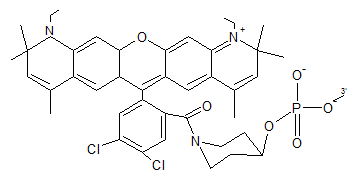
CFR 635 is an orange-red fluorophore. It is used as a good alternative to LC red 640. CFR 635 may be attached only to 5´end of an oligonucleotide.
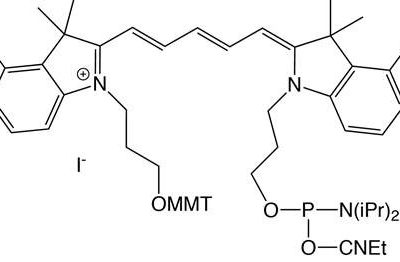
It is a deep red fluorophore. May be attached to the 5‘ end of the oligonucleotide. Oligonucleotide purification is a necessity.
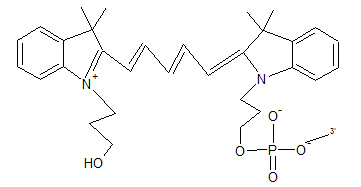
May be attached to the 5' end of the oligonucleotide. Oligonucleotide purification is a necessity.
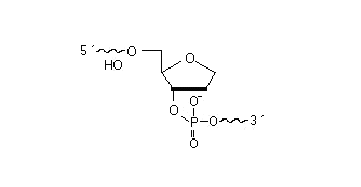
May be attached anywhere within the oligonucleotide sequence except for the 3' end. It is used to introduce a stable abasic position to the oligonucleotide chain.
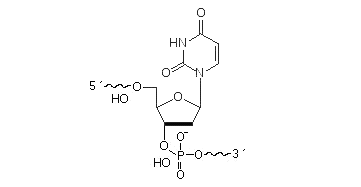
Deoxyuridine is usually introduced to the oligonucleotide DNA instead of deoxythimine. May be attached anywhere within the oligonucleotide sequence.
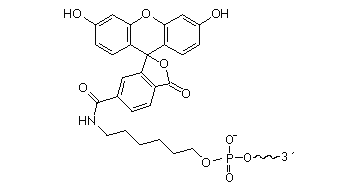
It is a mixture of 5- and 6-carboxyfluorescein isomers. Fluorescein may be attached anywhere within the oligonucleotide sequence.
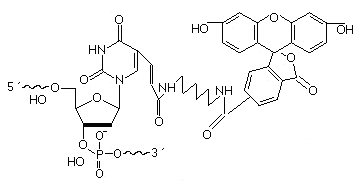
Fluorescein dT is attached by a six carbon spacer at position 5 of the thymine circle. Fluorescein dT may be attached anywhere within the oligonucleotide sequence.
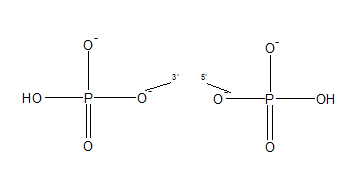
It can be linked to an oligonucleotide at the 5 'end and the 3' end. The 5 'end is used for ligation. The 3 'end serves as a 3' polymerase extension blocker.
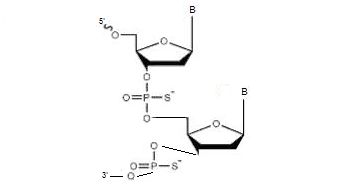
Phosphorothioates contain the exchange of one oxygen in the phosphate bond with sulfur. By exchanging oxygen for sulfur, the resistance of the oligonucleotide to nucleases increases.
Your request has been sent.
Your cycler has not been included in the list?
Send us the type of your cycler and we will find a solution for you.
Your request has been sent.
Thank you for your inquiry
Thank you for your inquiry
Your registration has been sent.
Thank you for your inquiry
Price will be calculated based on your exact location within 2 working days.