The ERMI software contains the solution for the entire process of sequencing – from sample preparation with the Sample Sheet Builder to the Variant Reporter that enables evaluation of sequencing data from patient samples. All in a user-friendly manner without the need for any prior coding or bioinformatic knowledge.
„ERMI Sample sheet builder“ allows you to create a sample sheet which is necessary for proper sequencing and generates a table with a combination of barcodes that can be used during library preparation of patient samples.
„ERMI Variant Reporter“ contains a bioinformatic pipeline that processes raw data from sequencer into annotated *.vcf and *.ermi files. Consequently, users can data browse and analyze in only a few clicks. ERMI software allows you to open individual analyzed patient samples with found variants categorized in four classes – confirmed, ambiguous, invalid, and rejected. Each found variant can be further opened, its coverage and other qualitative parameters browsed and also investigated in the connected Integrative Genomics Viewer[1] (IGV) program. Once the variant is defined as pathogenic, the user confirms it and includes it in the clinical sequencing report of the patient‘s sample. The clinical report is exported and additional information can be added. Moreover, users can create their own database of variants that are highlighted once confirmed in patients’ samples.
ERMI software is Java-based and compatible with macOS and Linux OS.
Minimal hardware requirements:
processor frequency 2 GHz; 4 cores; 16 GB RAM memory; storage capacity of hard-drive 1 TB
[1] Helga Thorvaldsdóttir, James T. Robinson, Jill P. Mesirov. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Briefings in Bioinformatics 14, 178-192 (2013).
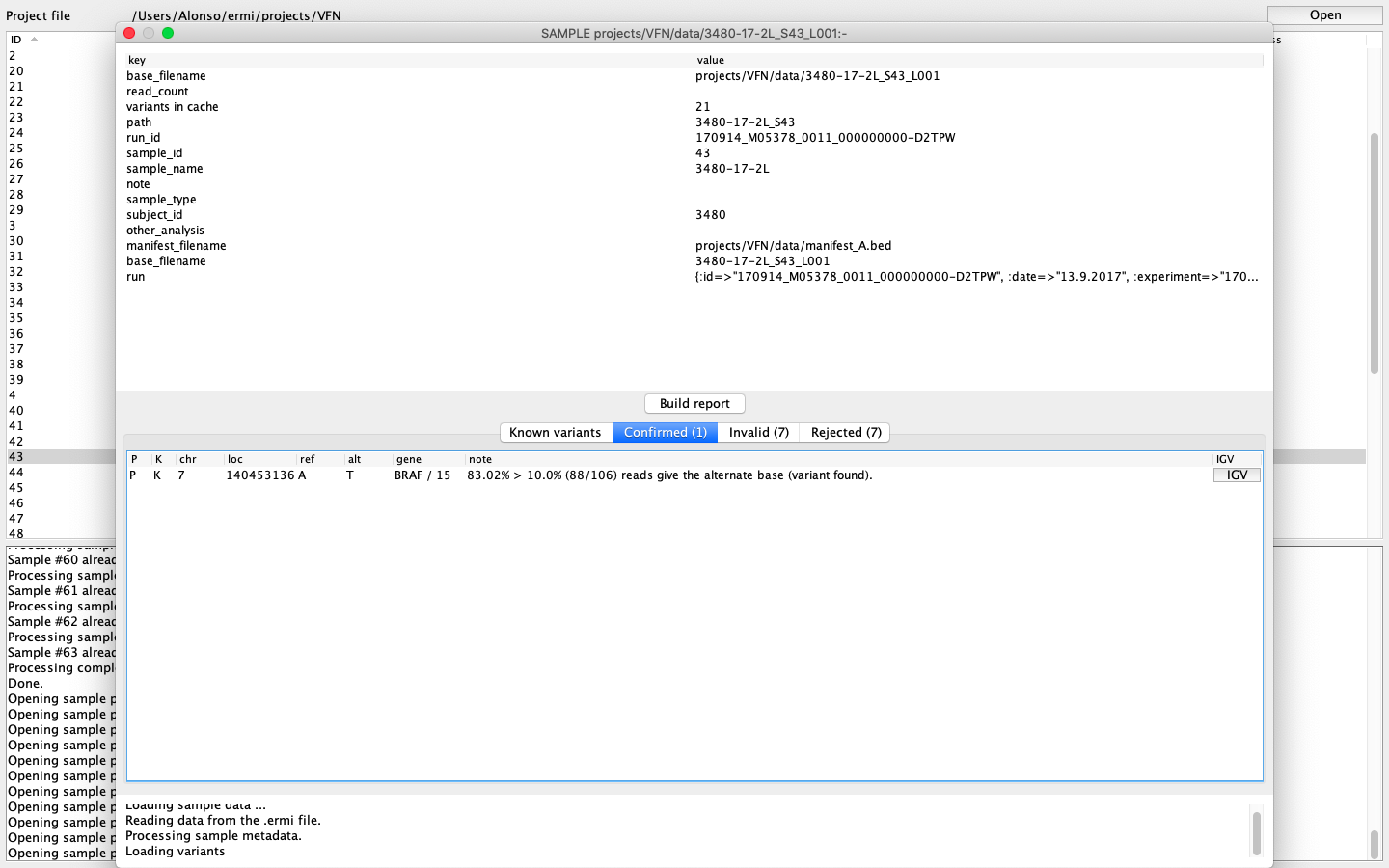
Fig.1: ERMI Variant Reporter – graphical interface of patient sample evaluation
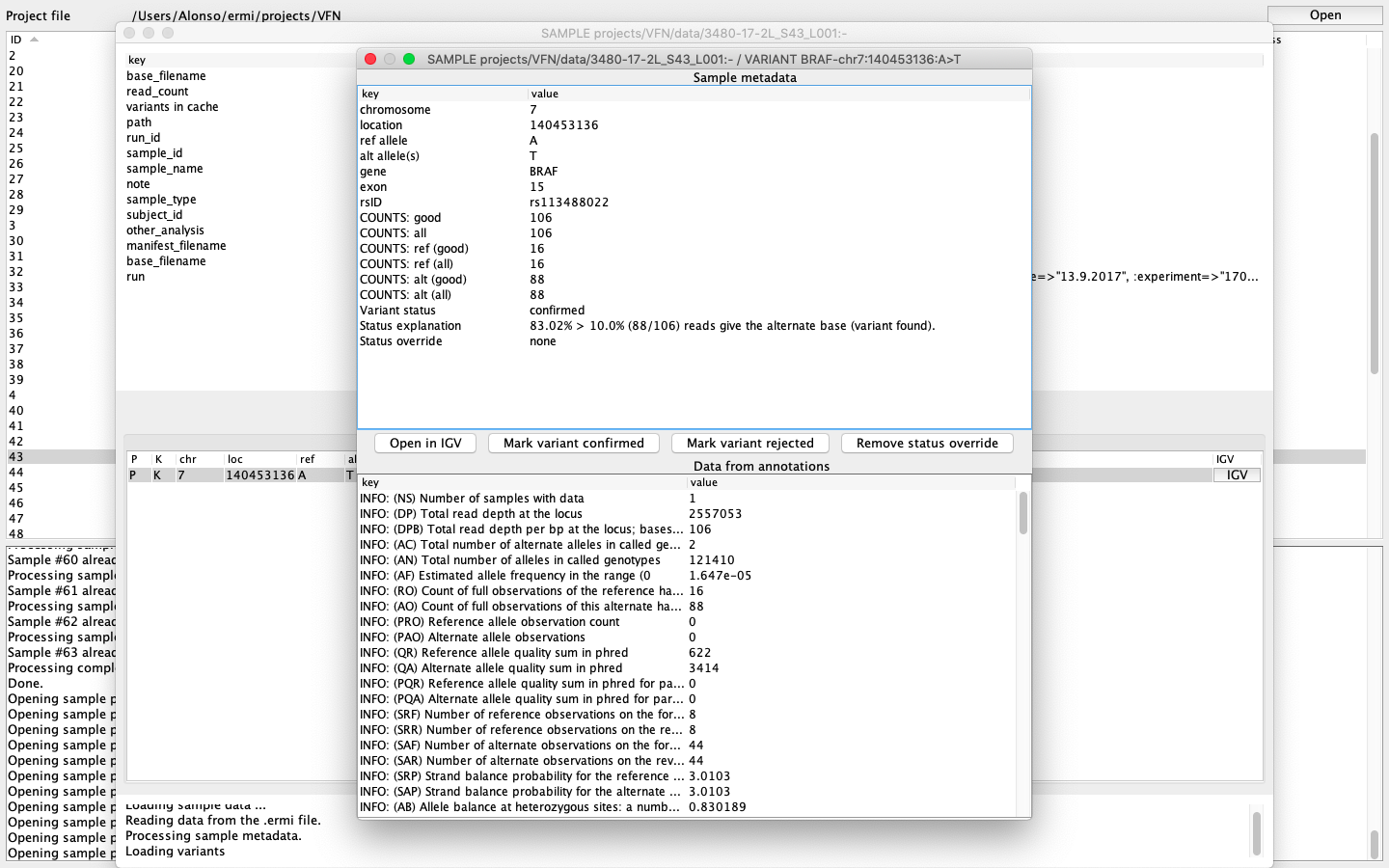
Fig.2: ERMI Variant Reporter – quality parameters of found variant
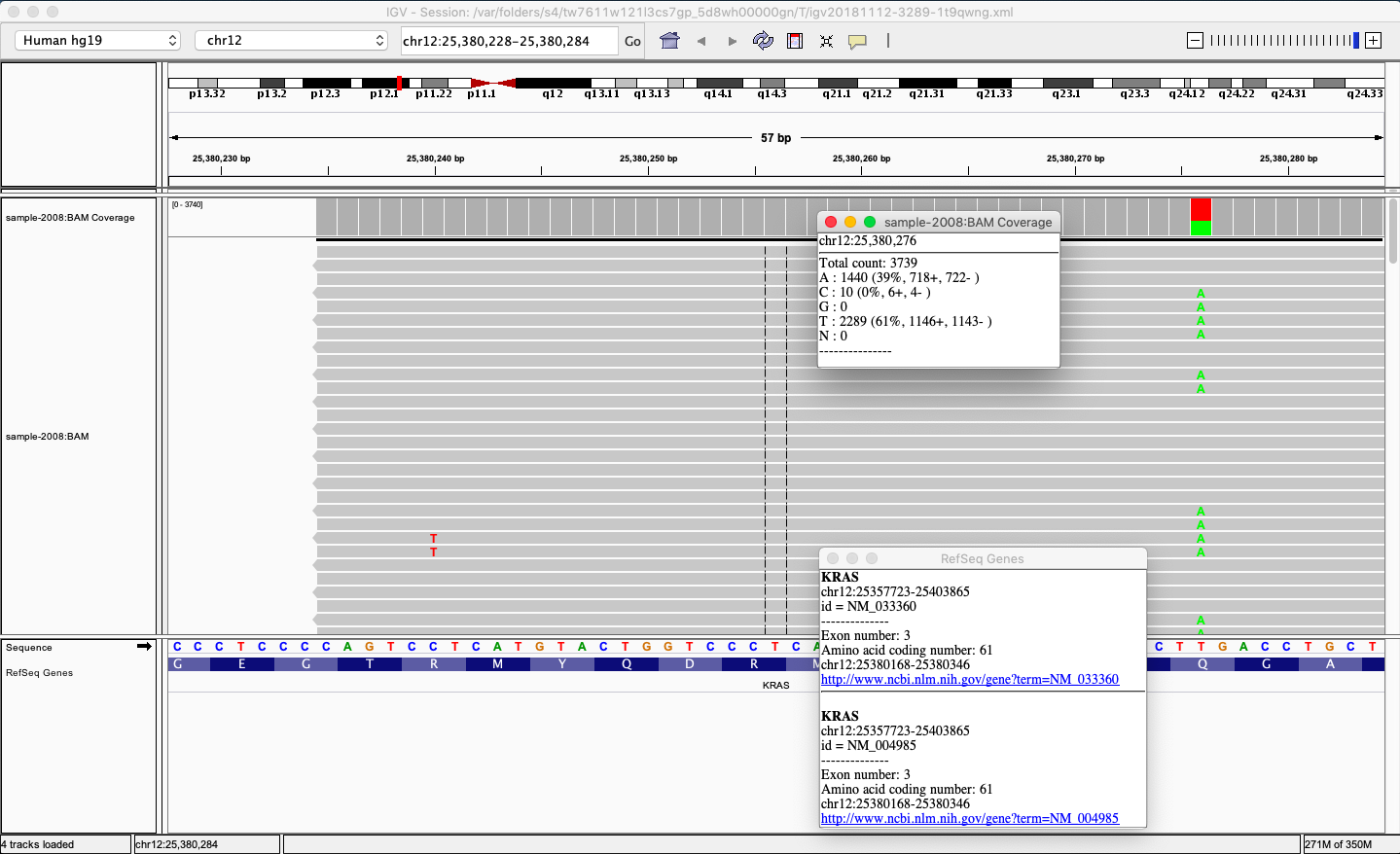
Fig.3: ERMI Variant Reporter enables direct connection with the IGV.